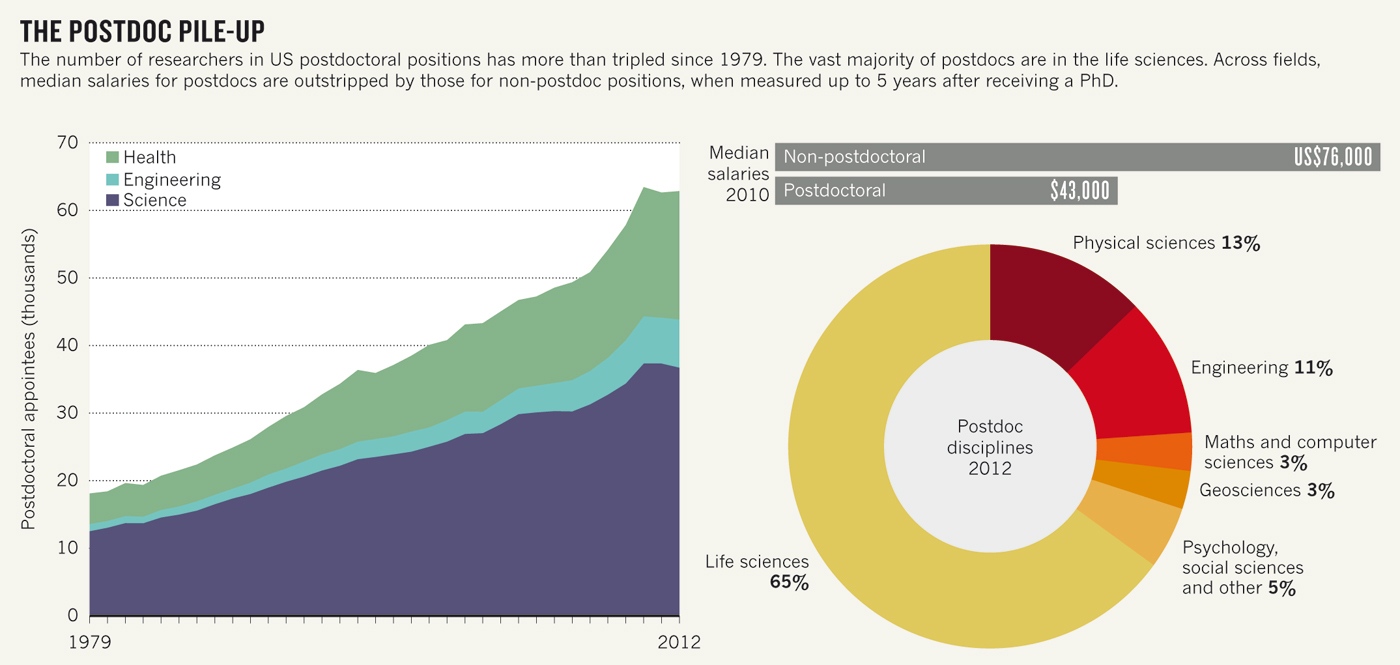
Structures of actin-like ParM filaments show architecture of plasmid-segregating spindles
Active segregation of
Escherichia coli low-copy-number plasmid R1 involves formation of a bipolar spindle made of left-handed double-helical actin-like ParM filaments
1, 2, 3, 4, 5, 6. ParR links the filaments with centromeric
parC plasmid DNA, while facilitating the addition of subunits to ParM filaments
3, 7, 8, 9. Growing ParMRC spindles push sister plasmids to the cell poles
9, 10. Here, using modern electron cryomicroscopy methods, we investigate the structures and arrangements of ParM filaments
in vitro
and in cells, revealing at near-atomic resolution how subunits and
filaments come together to produce the simplest known mitotic machinery.
To understand the mechanism of dynamic instability, we determine
structures of ParM filaments in different nucleotide states. The
structure of filaments bound to the ATP analogue AMPPNP is determined at
4.3 Å resolution and refined. The ParM filament structure shows strong
longitudinal interfaces and weaker lateral interactions. Also using
electron cryomicroscopy, we reconstruct ParM doublets forming
antiparallel spindles. Finally, with whole-cell electron cryotomography,
we show that doublets are abundant in bacterial cells containing
low-copy-number plasmids with the ParMRC locus, leading to an
asynchronous model of R1 plasmid segregation.
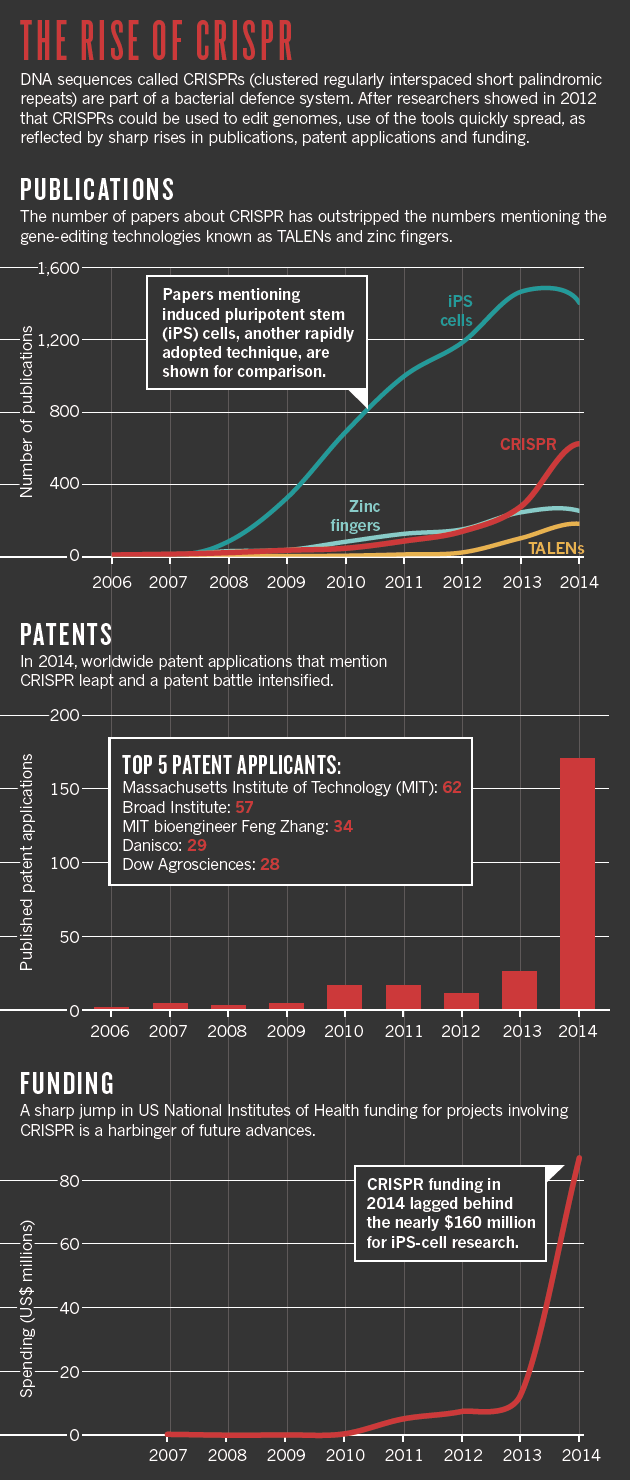
![ParM doublets in E. coli cells, imaged by cryo[hyphen]ET.](http://www.nature.com/nature/journal/vaop/ncurrent/images_article/nature14356-f4.jpg)
Fig.
a, A mutant of ParM that hydrolyses ATP more slowly
(D170A
) was overexpressed in
E. coli cells. Tomographic slices show large bundles of ParM blocking cell division. This experiment was performed two times.
b, The ParMRC operon driven from high
‐copy
‐number plasmid pDD19. Tomographic slice showing an example of observed doublets.
c, Tomographic slice for a medium
‐copy
‐number plasmid
(pKG321
).
d, Tomographic slice for a low
‐copy
‐number plasmid, emulating the native low
‐copy
‐number R1 plasmids
(pKG491, ‘mini
‐R1’ replicon
) in
E. coli (see
Supplementary Videos 5 and
6 to view entire tomograms
). Each experiment with different copy
‐number plasmids was performed once.
e,
Schematic depicting proposed asynchronous plasmid DNA segregation.
Bipolar ParM spindles are seeded when replication has produced two
parC
centromeric regions, still in close proximity. Each seeds one unipolar
ParM filament, which then come together in an antiparallel fashion to
form the segregating bipolar spindle. Non
‐productive
unipolar filaments or spindles that lack plasmid attachment will be
destroyed through the dynamic instability of ParM. This is in contrast
to earlier ideas in which all sister plasmids would be segregated
through one bundle of filaments, containing double the number of
unipolar filaments as the copy number of the plasmid in the cell
19.
Enlace al trabajo










![ParM doublets in E. coli cells, imaged by cryo[hyphen]ET.](http://www.nature.com/nature/journal/vaop/ncurrent/images_article/nature14356-f4.jpg)