Protein that delays cell division in bacteria may lead to the identification of new antibiotics
Scientists at Washington University have worked out how two bacterial strains delay cell division when food is abundant, an understanding that might be used to design drugs that stop division entirely
August 12, 2013
By Diana Lutz
LEVIN LAB
In a rapidly dividing chain of bacterial cells (top), constriction rings that will pinch the cells in two appear in red. The red doughnut to the bottom right of the image is a constriction ring seen head on rather than from the side. In the middle, an image of the constriction rings (red) has been overlaid on one of the cell walls (green), The bottom image shows the constriction rings (red) and the bacterial DNA (blue). Scientists at Washington University in St. Louis are learning exactly how the bacteria control the assembly of the constriction rings and thus the timing of cell division.
In 1958 a group of scientists working in Denmark made the striking observation that bacterial cells are about twice as large when they are cultured on a rich nutrient source than when they are cultured on a meager one. When they are shifted from a nutrient-poor environment to a nutrient-rich one, they bulk up until they have achieved a size more appropriate to their new growth conditions.
It has taken 60 years to figure out how the bacteria are able to sample their surroundings and alter their cell cycles so that they grow to a size suited to the environment.
In 2007 Petra Levin, PhD, a biologist at Washington University in St. Louis, reported in Cell that a soil bacterium named Bacillis subtilis has a protein that senses how much food is available and, when food is plentiful, temporarily blocks the assembly of a constriction ring that pinches a cell in two to create two daughter cells.
Now Norbert Hill, a graduate student in her group, reports in the July 25 online edition of PLoS Genetics that Escherichia coli uses a similar protein to help ensure cell size is coordinated with nutrient conditions.
Delaying division even just a little bit leads to an increase in daughter cell size. Once stabilized at the new size, cells take advantage of abundant nutrient sources to increase and multiply, doubling their population at regular intervals until the food is exhausted.
Because both the B. subtilis and E. coli proteins interact with essential components of the division machinery, understanding how they function will help in the discovery of antibiotics that block cell division permanently. A group in Cambridge, England, is already working to crystallize the E. coli protein docked on one of the essential components of the constriction ring.
If they are successful they may be able to see exactly how the protein interferes with the ring’s assembly. An antibiotic could then be designed that would use the same mechanism to prevent division entirely, killing the bacteria.
Why do bacteria get bigger on a good food source?
Bacteria increase and multiply by a process called binary fission. Each cell grows and then the divides in the middle to produce two daughter cells. What could be simpler?
But the closer you look, the less simple it becomes. For binary fission to work the cell must make a copy of its circular chromosome, unlink and separate the two chromosomes to create a gap between them, assemble a constriction ring in the middle of the cell and coordinate the growth of new cell membrane as the ring cinches tight and pinches the mother cell in two. To complicate matters, bacteria don’t necessarily do these steps one by one but can instead work on several steps simultaneously.
Most of the time the goal is to produce daughters the same size as the mother cell. But when food is plentiful, bacteria start making more copies of their DNA (as many as 12) in anticipation of divisions to come, and they can’t easily cram all the extra DNA into standard-sized cells. So they grow bigger to accommodate the extra genetic material and remain large as long as the food lasts.
The inventory of partly copied chromosomes fuels rapid population growth, because a cell doesn’t start from scratch when it needs another copy of its chromosome. Under optimum conditions, E. coli, for example, divides once every 17 minutes. If they are allowed to grow unhindered this means that in 24 hours 1 bacterium becomes about 5 x 1021 bacteria (that is 5 with 21 zeros after it.)
How do bacteria know the pickings are rich?
In B. subtilis and E. coli the signal is a modified sugar called UDP-glucose. Presumably, the richer the growth medium, the higher the level of this sugar inside the cell.
Norbert Hill (“Bisco”), the first author of the PlosGenetics paper, in the lab. He worked out the signaling pathway in E. coli that connects nutrient levels to cell division, largely by studying mutant strains of E. coli with broken pathways.
In both bacteria UDP-glucose binds to a protein and the sugar-protein complex then interferes with the assembly of the constriction ring. In the case of B. subtilis the protein is called UgtP and in the case of E. coli it is OpgH.
“It’s interesting,” Hill said, “that both organisms, which are more different from one another than we are from bakers’ yeast, are using the same system to coordinate changing size in response to nutrient availability.”
UgtP and OpgH are bifunctional proteins that are “moonlighting” as elements of the cell-division control systems. In both cases their day jobs are to help build the cell envelope. “We think they are communicating not only how much glucose there is in the cell, but also how fast the cell is growing,” Levin said. “The sensor says not only is food abundant, but we’re also growing really fast, so we should be bigger.”
Both proteins delay division by interfering with FtsZ, the first protein to move to the division site, where it assembles into a scaffold and recruits other proteins to form a constriction ring.
“Very little is known about the assembly of the ring,” Hill said. “There are a dozen essential division proteins and we don’t know what half of them do. Nor do we understand how the ring develops enough force to constrict.”
“We do know FtsZ exists in two states,” Hill added. “One is a small monomer and the other is many monomers linked together to form a multi-unit polymer. We think the polymers bind laterally to form a scaffold and then, with the help of other proteins, make a meshwork that goes around the cell.
UgtP and OpgH both interfere with the ability of FtsZ to form the longer polymers necessary for assembly of the constriction ring.
When nutrient levels are low, UgtP and OpgH are sequestered away from the division machinery. FtsZ is then free to assemble into the scaffold supporting the constriction ring so the cell can divide. Because division proceeds unimpeded, cells are smaller when they divide.
What about other bacteria?
This control system helps to explain the 60-year-old observation that bacterial cells get bigger when they are shifted to a nutrient-rich medium.
Comparing the mechanisms that govern cell division in E. coli and B. subtilis reveals conserved aspects of cell size control, including the use of UDP-glucose, a molecule common to all domains of life, as a proxy for nutrient availability, and the use of moonlighting proteins to couple growth-rate-dependent phenomena to the central metabolism.
But much more is known about these model organisms, which many labs study, than the average bacterium. Nobody is sure how many species of bacteria there are—somewhere between 10 million and a billion at a guess—and they don’t all divide the way B. subtilis and E. coli do.
The whimsically named giant bacterium Epulopiscium fiselsoni (“Fishelson’s guest at a fish’s banquet”) that lives in the guts of sturgeonfish, has the gene for FtsZ but doesn’t divide by binary fission. And then there are bacteria like the pathogen Chlamydia traachomatis that don’t have a gene for anything like FtsZ. “We don’t know how these bacteria divide, much less maintain an appropriate cell size,” Levin said.
Reloj
martes, 13 de agosto de 2013
lunes, 5 de agosto de 2013
The End of Antibiotics? - Body Horrors | DiscoverMagazine.com
The End of Antibiotics?
By Rebecca Kreston | August 1, 2013 7:00 pm
Maryn McKenna has an unsettling and sobering article at Nature examining the the emergence of carbapenem-resistant Enterobacteriaceae. Since 2002, this large family of bacteria, gram-negative organisms that include many symbionts as well as the gut-dwelling Escherica coli and Klebsiella species that cause hospital infections, are increasingly in possession of a carbapenem-resistance gene rending our best antibiotics useless.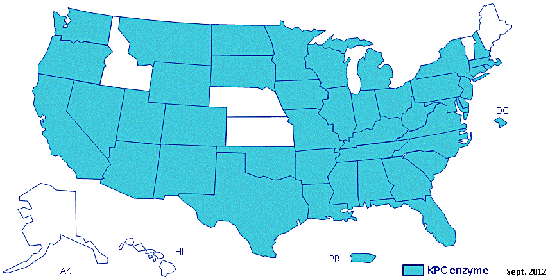
A map of the United States showing states with carbapenemase-producing CRE that promote resistance to carbapenem antibiotics as confirmed by CDC as of September 2012.

A graph from the Center for Disease Dynamics, Economics & Policy. Isolates of cabapenem-resistant Klebsiella pneumoniae have emerged since the early 2000s and are becoming increasingly common throughout the country with resistance rates rising from 0% until 2002 to 4.5% in 2010. K. pneumoniae, a member of the Enterobacteriaceae family, causes the vast majority of hospital-borne urinary tract infections (UTIs) and bloodstream infections. Click for source and to see an interactive version of this image.
In 2011, the CDC released a fact sheet for the public, “Antibiotics: Will they work when you really need them?” to help promote responsible antibiotic usage. In a decade, will we even be able to ask this question?
Resources
The World Economic Forum releases an annual survey, the Global Risk report, on the top 50 risks facing our world. Their 2013 report included antibiotic-resistant bacteria as one of those risks, citing destabilization to our health systems and food supply, imperiling the practice of common medical procedures, and the fact that none of the antibiotics and drugs that are currently in the development pipeline would be able to protect us against certain bacteria. Must read.
The End of Antibiotics? - Body Horrors | DiscoverMagazine.com
sábado, 3 de agosto de 2013
Suscribirse a:
Comentarios (Atom)
